Online-Kolloquium (Link zur Veranstaltung auf Zoom, Meeting ID: 876 5905 1997, Passcode: 780903)
The mechanical properties of cells have long been established as a sensitive and label-free biomarker for cell and tissue function. While mechanical cell assays have traditionally been limited to low throughput or small sample size, our development of real-time deformability cytometry increased analysis rates to up to 1,000 cells per second [1,2] . In my presentation I will discuss how this technology provided a new perspective on life science research by enabling the first time a high-throughput mechanical characterization of heterogeneous biological samples on a single cell level. This includes monitoring the invasion of the Malaria parasite into erythrocytes, studying the impact of magnetic nanoparticles on blood platelets, and to assess the quality of manufactured red blood cells [3-5].
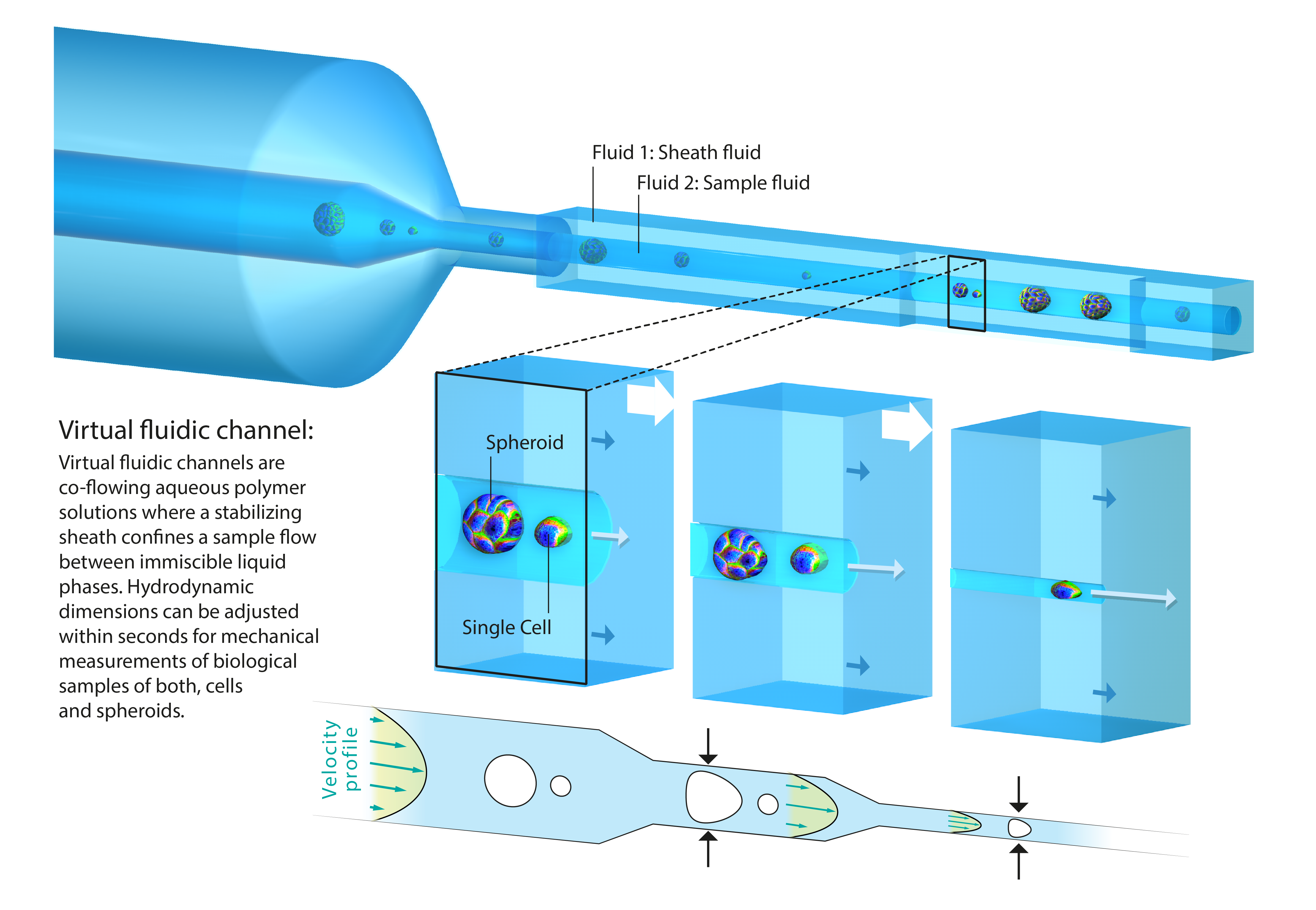
The second part of my presentation focuses on multicellular structures and the question how mechanical properties of cells contribute to tissue rheology. Until recently, a high-throughput technology for mechanical characterization of tissue had been elusive. We answered this challenge by introducing virtual fluidic channels [6] . Virtual channels are liquid-bound microfluidic constrictions of variable cross-sections, which can be modified within seconds and tailored to hydrodynamic stress distributions to match a wide size range of biological samples. We perform high-throughput rheology on spheroids as a 3D tissue model system and demonstrate that the Young’s modulus of isolated cells exceeds the one of tissue by one order of magnitude. Interestingly, our results suggest that tissue elasticity is mainly driven by cell-cell interactions and extracellular matrix contributions.
References:
- O. Otto, P. Rosendahl, A. Mietke, S. Golfier, C. Herold, D. Klaue, S. Girardo, S. Pagliara, A. Ekpenyong, A. Jacobi, M. Wobus, N. Töpfner, U.F. Keyser, J. Mansfeld, E. Fischer-Friedrich, and J. Guck
Real-time deformability cytometry: on-the-fly cell mechanical phenotyping Nature Methods 12, 199-202 (2015) - B. Fregin, F. Czwerwinski, D. Biedenweg, Girardo, S., Gross S., K. Aurich, and O. Otto
Dynamic real-time deformability cytometry: high-throughput single-cell rheology in complex samples Nature Communications 10, 415 (2019) - M. Koch, K.E. Wright, O. Otto, M. Herbig, N.D. Salinas, N.H. Tolia, T.J. Satchwell, J. Guck, N.J. Brooks, and J. Baum
Plasmodium falciparum erythrocyte-binding antigen 175 triggers a biophysical change in the red blood cell that facilitates invasion PNAS 114, 201602843 (2017) - K. Aurich, B. Fregin, R. Palankar, J. Wesche, O. Hartwich, D. Biedenweg, TH. Nguyen, A. Greinacher, and O. Otto
Label-free on chip quality assessment of cellular blood products using real-time deformability cytometry Lab on a Chip (accepted for publication) - E. Guzniczak, O. Otto, G. Whyte, N. Willoughby, M. Jiminez, and H. Bridle
Deformability-induced lift force in spiral microchannels for cell separation Lab on a Chip 10.1039/c9lc01000a (2020) - M.H. Panhwar, F. Czerwinski, V.A.S. Dabbiru, Y. Komaragiri, B. Fregin, D. Biedenweg, P. Nestler, R.H. Pires, and O. Otto
High-throughput cell and spheroid mechanics in virtual fluidic channels Nature Communications 11, 2190 (2020)