Project C4
Doctoral researcher: Daniel Troitsch
Principle investigator: Susanne Sievers
Co-supervisors: N.N
The membrane of the intestinal pathogen C. difficile as the first line of defense during infection-related oxygen and bile acids challenge
The strict anaerobic bacterium Clostridium difficile has become the most frequent cause of bacterial nosocomial infections [1]. The pathogen can thrive in our gut when the intestinal microbiome is suppressed. C. difficile toxins cause inflammation of the intestinal epithelium resulting initially in diarrhea, but in advanced infections in pseudomembranous colitis (PMC), toxic megacolon, bowel perforation and sepsis. The bacterium can form endospores which will survive antibiotics treatment explaining the high relapse rate of the infectious disease. Thus, novel antimicrobial strategies are needed to ultimately cure people from C. difficile infections. This project aims at identifying structures and mechanisms in the bacterial membrane that quench the destructive action of bile acids and traces of oxygen, both represent stress factors the pathogen is challenged with in our intestines.
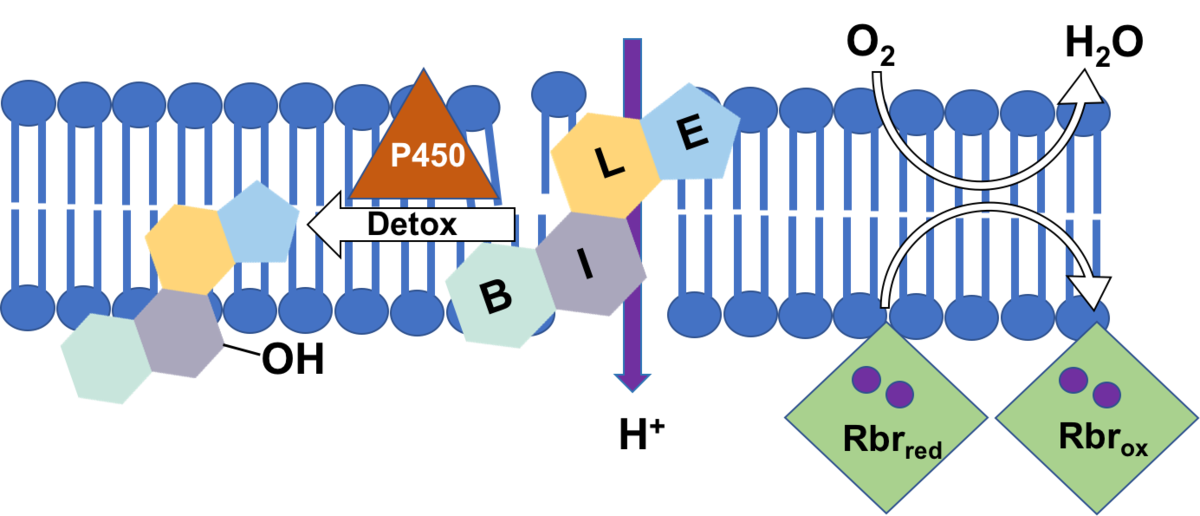
Specifically, we will investigate if rubrerythrins are involved in the detoxification of oxygen in C. difficile as was shown for several other anaerobic bacteria [2]. Rubrerythrins with their non-heme diiron centre reduce oxygen to water, and their membrane location is conceivable [3]. C. difficile encodes four rubrerythrins in its genome, three of them were found to be expressed in late exponential phase [4]. First growth experiments showed a high tolerance of C. difficile towards oxygen. Since one of the expressed rubrerythrins represents the second most abundant protein in C. difficile, we will test for its ability to scavenge oxygen with partners of project B4 (Kahlert & Lalk) and benefit from the expertise in iron-sulfur cluster proteins of investigators of project A1 (Lillig). Profiting from the metabolomics expertise in project B4 we also want to characterize oxidative changes in the membrane lipid pattern of C. difficile after oxygen exposure, in wildtype cells and in mutant strains missing one or several of the rubrerythrins.
Bile acids represent amphiphilic substances challenging the integrity of the bacterial cell. First data show that C. difficile can adapt to high concentrations of bile acids which is probably accompanied with a restructuring of the bacterial cell envelope. Preliminary data revealed an increased expression of several subunits of ATP synthase when cells are stressed with bile acids. Since C. difficile ferments amino acids for its energy supply and does not use an electron transport chain to build up a proton gradient across the membrane, we assume that the increased ATP synthase is needed to pump protons in order to keep the membrane potential.
Employing fluorescent dyes and making use of the expertise of partners of project B1 (Helm) and B2 (Scholz) we would like to investigate the depolarizing effect of different bile acids on membranes. Bile acids feature a steroid structure, thus representing toxic compounds which can only be attacked and degraded by special enzymes as P450 monooxygenases for instance. In cooperation with experts of project A2 (Bornscheuer) we will look for enzymes in C. difficile that could modify/degrade bile acids.
Finally, we want to explore the infection-simulating condition of concomitant bile acids and oxygen challenge to prove the assumption that cell envelope distortion through bile acids will result in increased sensitivity to oxygen.
We feature a strong expertise in membrane proteomics and redox proteomics which will be supportive to partners of project A1 (Lillig) and C3 (Stöhr).
Literature
[1] Robert Koch-Institut: Infektionsepidemiologisches Jahrbuch 2012 - 2015 Berlin
[2] Hillmann F, Riebe O, Fischer R-J, Mot A, Caranto JD, Kurtz DM Jr, & Bahl H. (2009) Reductive dioxygen scavenging by flavo-diiron proteins of Clostridium acetobutylicum. FEBS Letters 583:241–245.
[3] Shanklin J, Achim C, Schmidt H, Fox BG, & Münck E. (1997) Mössbauer studies of alkane omega-hydroxylase: evidence for a diiron cluster in an integral-membrane enzyme. PNAS 94:2981–2986.
[4] Otto A, Maass S, Lassek C, Becher D, Hecker M, Riedel K, & Sievers S. (2016) The protein inventory of Clostridium difficile grown in complex and minimal medium. Proteomics Clin Appl. 10:1068-1072.
Contact
Susanne Sievers
University of Greifswald
Institute of Microbiology
Felix-Hausdorff-Str. 8
D-17487 Greifswald
Tel: +49 (0)3834 420 5915
Fax:+49 (0)3834 420 5902
sieverss(at)uni-greifswald(dot)de
Website